Software
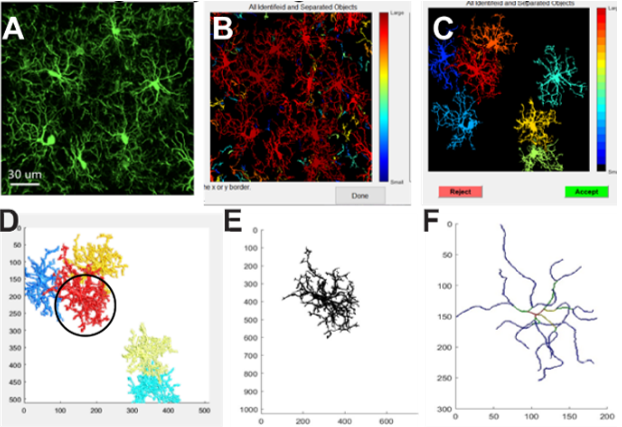
CellSelect-3DMorph reconstructs cells from fluorescent pixel data, providing key parameters such as cell volume, territorial volume, ramification index, branch length, and the number of branches and endpoints. It builds on the 3D Morph code, adding the ability to exclude incomplete cells from analysis, ensuring accurate morphological calculations.
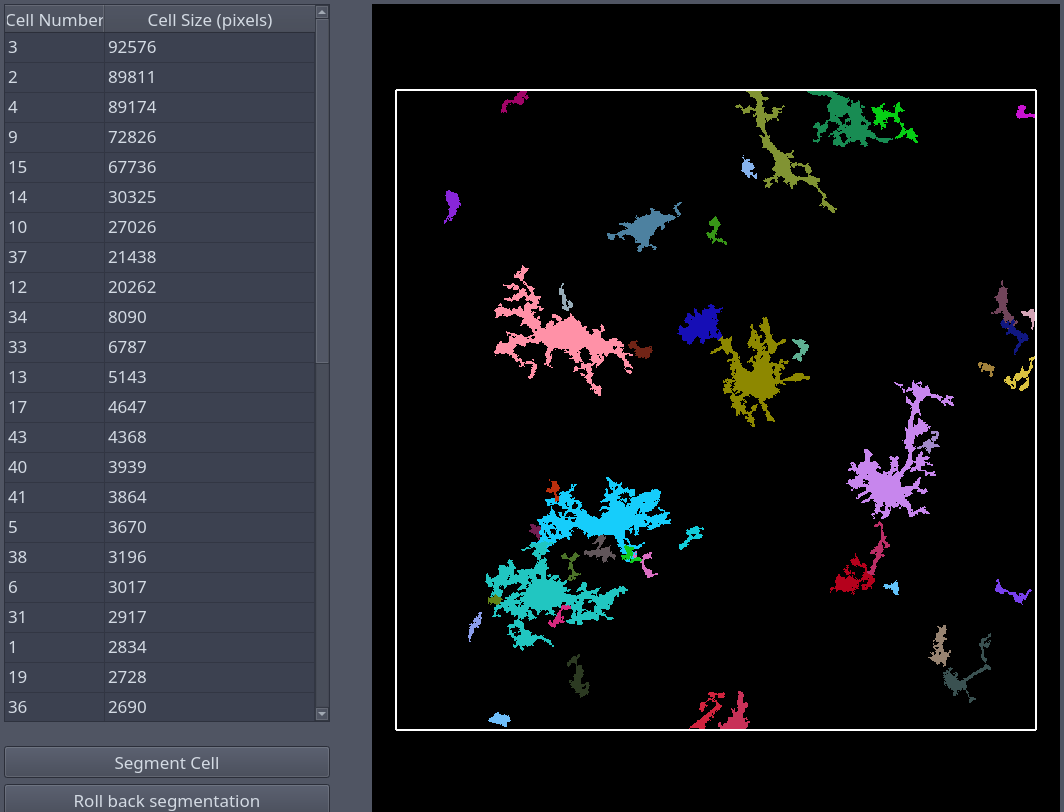
Pycroglia is a modern, open-source Python port of CellSelect-3DMorph that enables quantitative 3D microglia morphology analysis. By reconstructing individual cells voxel by voxel, it extracts morphological descriptors such as cell volume, territorial volume, ramification index, branch length, and number of branches and endpoints. It supports both GUI and library modes for interactive and automated workflows.
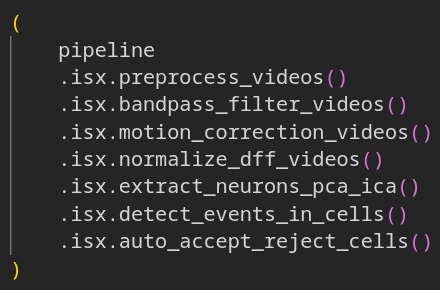
CIPipeline is a Python library for building and running reproducible, library-agnostic calcium-imaging processing pipelines. It provides adapters for Inscopix and CaImAn, utilities, plotters and example Jupyter notebooks.
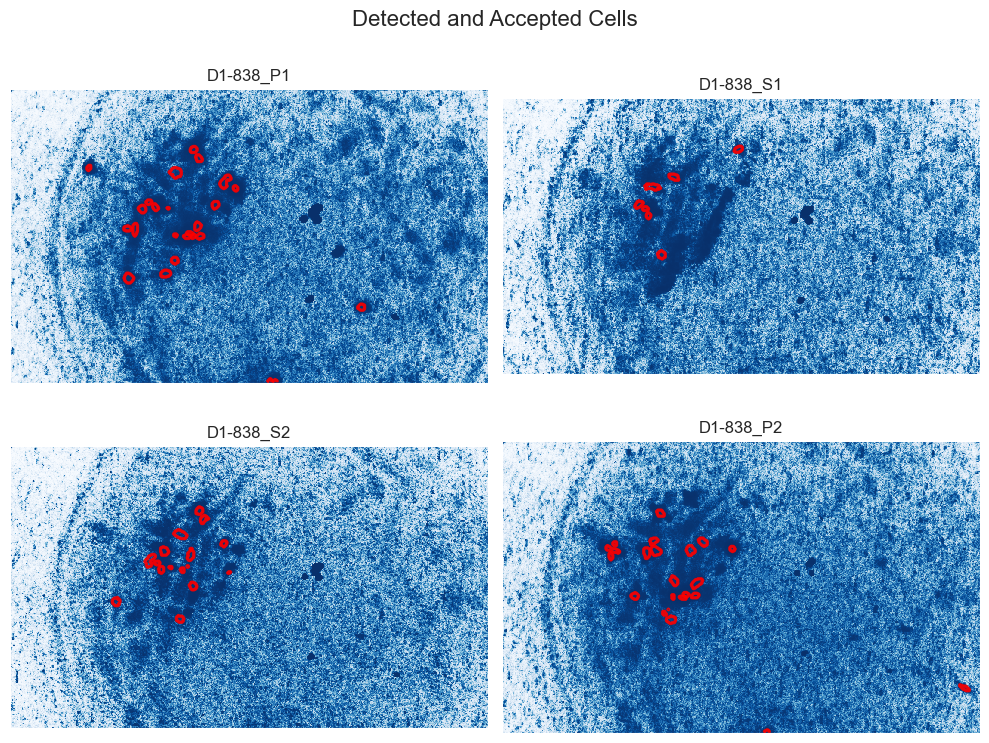
This pipeline reduces the space required to analyze calcium imaging data using the Inscopix Data Processing algorithms, allowing users to easily remove intermediate steps while leaving metadata to recompute them if necessary. It combines multiple functions used for cell extraction in a simple method call and can process multiple recordings together.